Body parts (e.g. face, limbs, trunk) are made of composite tissues, which are organized in assembled tissue layers (e.g. skin, fat and cartilage for the ear) with an intertwining vascular and nervous network. Each of the layers is structured by its extracellular matrix (ECM) that provides physical and biochemical cellular support. Tissue Engineering (TE) of composite tissues is a subfield of Regenerative Medicine, in which regeneration of a damaged body part is envisioned through the manufacturing of TE constructs: i.e. ECM-based or synthetic scaffolds seeded with cells and/or growth factors. ECM-based TE constructs can currently be derived from decellularized composite tissues (animal- or human-derived) as a scaffold and a subsequent recellularization process with new cells, but this process still needs further optimization. Synthetic scaffolds would foster the development of TE constructs, but remain a hypothetic option due to the lack of knowledge on the required building blocks to mimic the native 3D morphology and organization of the different ECM layers, and hence the ideal synthetic TE construct for regeneration of composite tissues is still far from being reality.
The aim of the project is thus to improve the knowledge on the required building blocks of the different layers within the extracellular matrix constituting the composite tissues in order to promote the manufacturing of synthetic tissue-engineering constructs.
To achieve this goal, the project contains three sub-objectives:
1) Optimizing the creation of ECM-based scaffolds using perfusion-decellularization techniques
2) Optimizing the 3D anatomical characterization based on microCT and CECT
3) Developing artificial intelligence-based image processing using deep-learning and CNN
In the frame of this project, bio-blueprints stands for a highly detailed, statistically relevant quantitative description of the “full 3D” morphology of the different extracellular matrix types/layers and the intertwining vascular/nervous network within composite tissues
Perfusion-decellularization
Currently existing TE strategies still suffer from unpredictable and qualitatively inferior results, thus hampering clinical translation. This low repeatability or lack of robustness is often caused by widely variable scaffold properties (due to inconsistency in the design process). Hence, the optimal scaffold structure is still under debate. Material-specific variability should be reduced and trial-and-error should be avoided. Therefore, the aim of Bio-blueprints is initiate a paradigm shift in composite tissue TE by using nature’s own bio-blueprints for a more intelligent design of both ECM-based and synthetic TE constructs. Using this approach, we aim at an increase in repeatability and robustness in the final outcome, i.e. the regeneration of the composite tissues in body parts.
Indeed, in order to be the most likely to get to clinical applications, we need to produce scaffolds that can appropriately be repopulated by a recipient’s cells, either during in vitro (using bioreactor) or in vivo (after replantation) maturation. These scaffolds will then need to fulfil two main things: they need to mimic the native structural complexity and they have to have a patent and accessible vascular tree. Complexity is mandatory to provide cells a correct environment for attachment, proliferation and differentiation towards the right tissue type. Synthetic matrices are for the moment lacking both aspects, explaining the current limited clinical applications of the field.
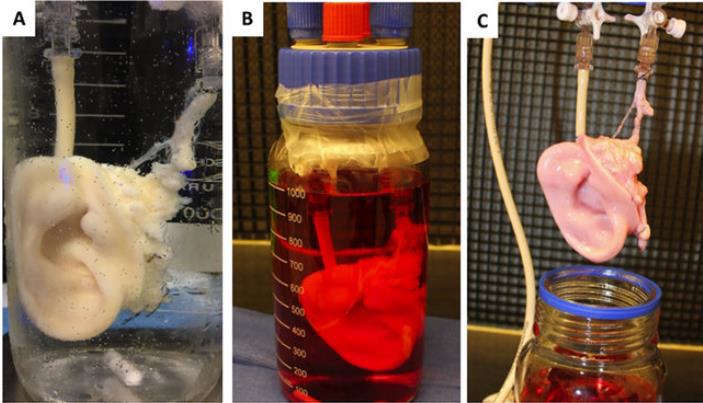
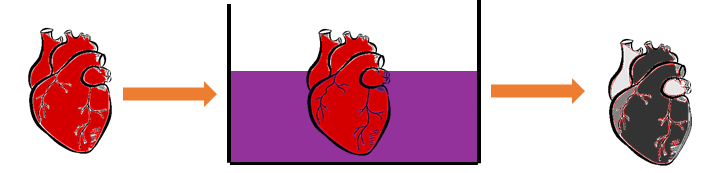
Active perfusion is of tremendous importance to ensure survival, through a vascular network, of seeded cells in the construct; it should then be reconnected to the host own circulation, like for any organ or composite tissue allotransplant. The “perfusion-decellularization” technique, may represent the ‘holy grail’ of immunosuppression-free transplantation. The basics of this technique involve circulating a mild detergent through a harvested organ/tissue via the native vascular system with the goal of removing cellular components, thus creating an ECM-based scaffold retaining original tissue architecture and properties (= decellularization procedure). The ECM scaffold could then be repopulated with autologous cells that would lead to a biological identity reassignment and immune tolerance once implanted, see example in Figure 1.
Contrast-enhanced Computed Tomography
The gold standard technique for structural analysis of biological tissues is currently classical 2D histology. In this process, the tissue is fixed, sliced in very thin sections (usually 5 µm thick) and stained. This results in a stack of 2D microscope images, representing the 3D arrangement of the sample. While the individual slices can be of very high quality (highly selective stains, high in-plane spatial and contrast resolution), the resolution along the third dimension in space is often limited due to the restricted amount of sections that can be realistically sliced and imaged for one sample (Figure 2). These drawbacks penalize the spatial assessment of highly heterogeneous tissues (e.g. composite tissues), including the quantification of the spatial relationship between the different ECM types/layers and the intertwining vascular network within one sample.
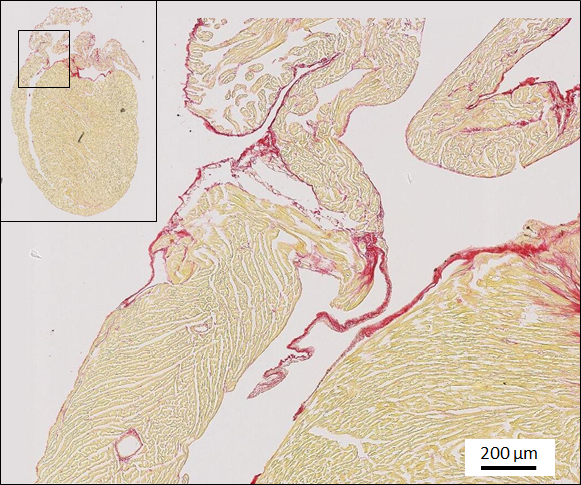
Figure 2 : in-plane view of a 2D classical histology section of a mouse heart after picrosirius red staining (left) and 3D stack of the 2D sections illustrating the loos of information (right).
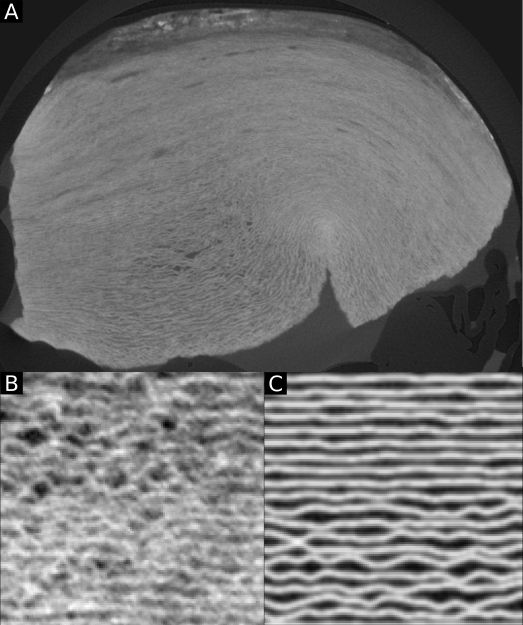
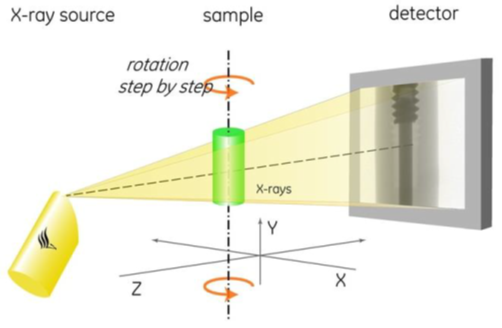
To counteract these drawbacks, ex vivo high-resolution microfocus computed tomography (microCT) and contrast-enhanced microCT (CECT) imaging are used to obtain a full 3D visualization of the samples. In microCT imaging, the sample is placed on a rotating stage and is penetrated by X-rays at several angles over 360°. Projections are received by the detector and are then reconstructed to form a 3D volume (Figure 3, top). While X-ray-based imaging techniques are known to be powerful to image mineralized biological tissues, such as bone, the visualization of soft tissues requires the addition of a contrast-enhancing staining agent (CESA), resulting in CECT. CESAs are chemical compounds that interact with the tissue through passive diffusion and render it X-ray attenuating (Figure 3, bottom). In this project, Hafnium-substituted Wells-Dawson polyoxometalate (Hf-WD POM) is used as CESA to stain the samples without inducing tissue shrinkage or dehydration.
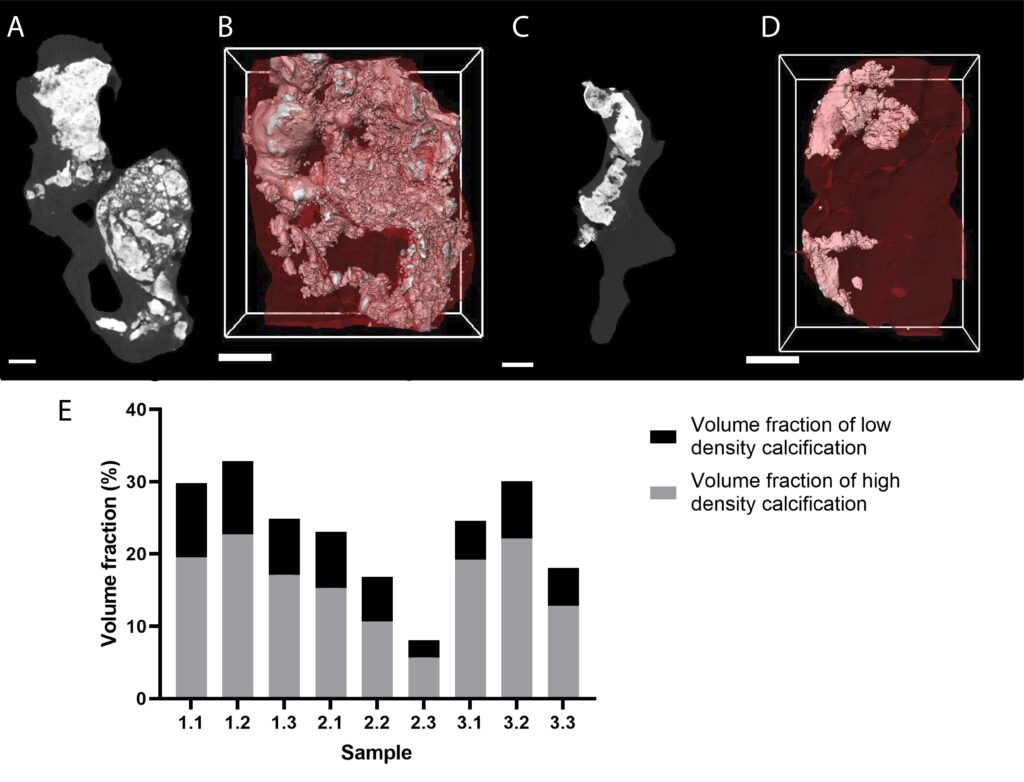
In this project, microCT and CECT are optimized to characterize the microstructure of heart and more specifically of heart valves. We started by reviewing the ex vivo characterization of the cardiovascular system using X-ray (reference to publication). In our experiments, microCT is used without contrast enhancement to characterize the calcifications that grow in the middle of the soft tissue, for instance in case of aortic stenosis. It allows a quantitative characterization of the calcifications at high resolution (Figure 4). Moreover, although microCT has never been used to assess the microstructural components of the valve (collagen, elastin, proteoglycans), we are convinced that CECT is a highly valuable tool to study the microstructure of heart valves in healthy and diseased conditions but also after decellularization.
Deep Learning-based tissue segmentation and characterization
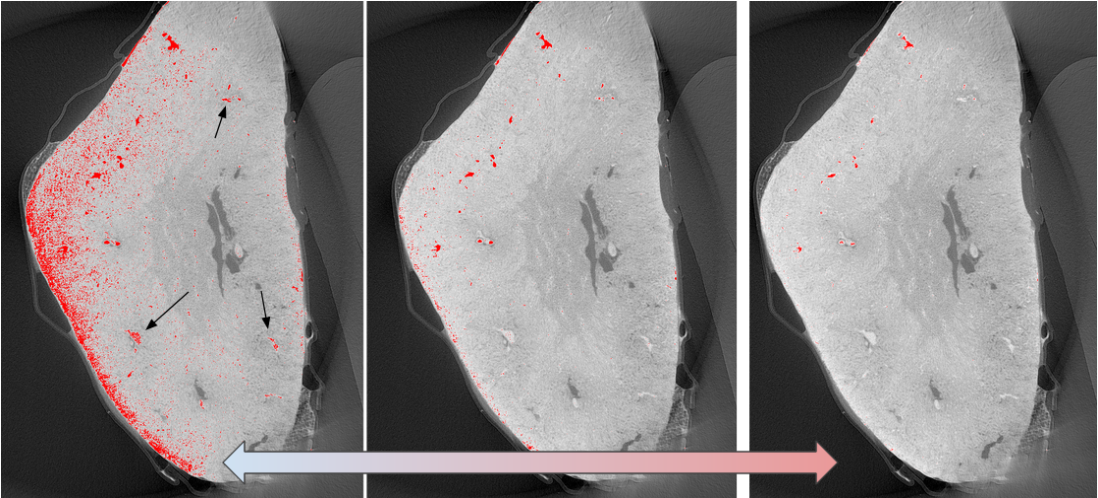
Bio-blueprints has the objective to design and train 3D image analysis CNN models that are robust and/or adaptable to input image variability, despite the lack of a large and representative set of paired image/segmentation samples.
Recent methods have been proposed to compensate the lack of annotated samples, but they are designed for large datasets of (natural) color images (or videos), while our approach will need to work with much smaller datasets from a completely different modality, making pre-training untrustworthy. We therefore consider the following strategies :
- Self-supervised training strategies, which can exploit images without labels;
- Exploiting prior knowledge through regularization of traditional semi- and unsupervised methods and label-augmentation during training. (Figure 5)
Such techniques are applicable when the ground-truth labels exhibit internal structure. In Bio-blueprints, some datasets exhibit region connectivity (in case of cartilage, muscle, adipose tissue, tendon, fibers, etc.) and others ramifications (in case of blood vessels, nerves, fibers, etc.).